Implementation in in Matlab, Python and in NeuroML2/LEMS of Jorge F. Mejias, John D. Murray, Henry Kennedy, and Xiao-Jing Wang, “Feedforward and Feedback Frequency-Dependent Interactions in a Large-Scale Laminar Network of the Primate Cortex.” Science Advances, 2016 (bioRxiv).
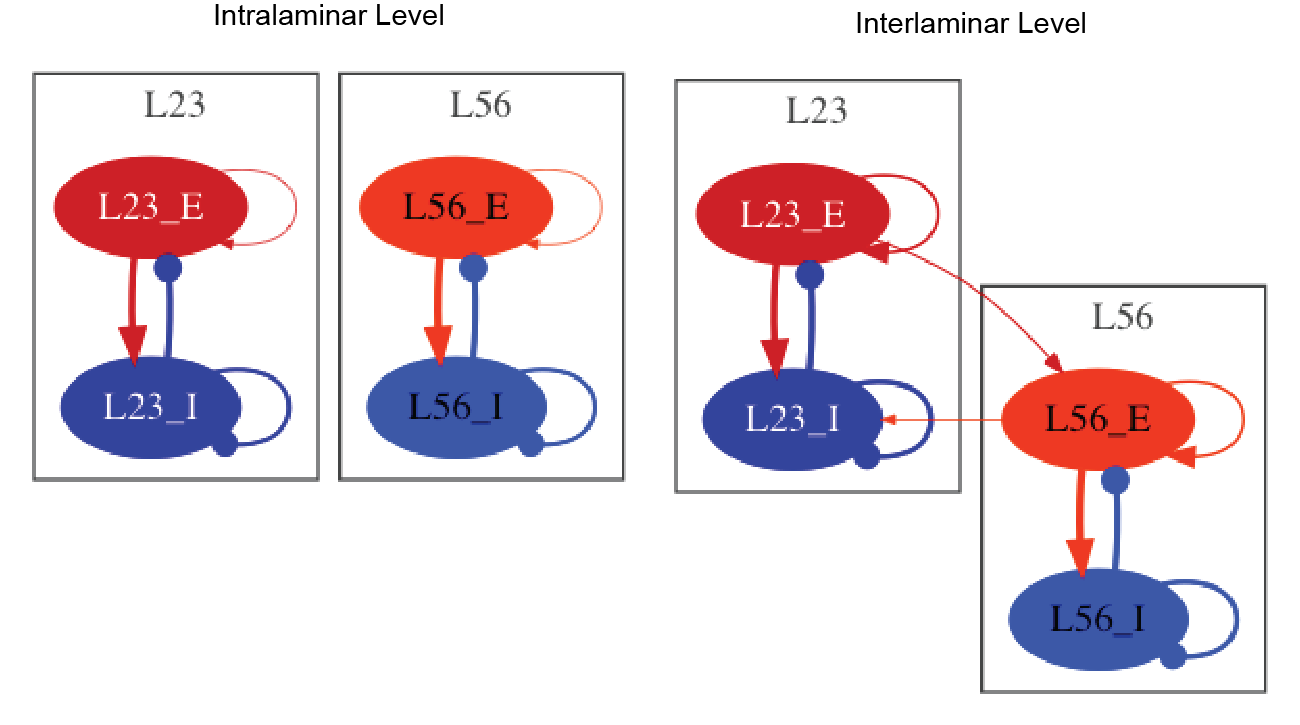
The model simulates the dynamics of a cortical laminar structure across multiple scales: (I) intralaminar, (II) interlaminar, (III) interareal, (IV) whole cortex. Interestingly, the authors show that while feedforward pathways are associated with gamma oscillations (30 - 70 Hz), feedback pathways are modulated by alpha/low beta oscillations (8 - 15 Hz).
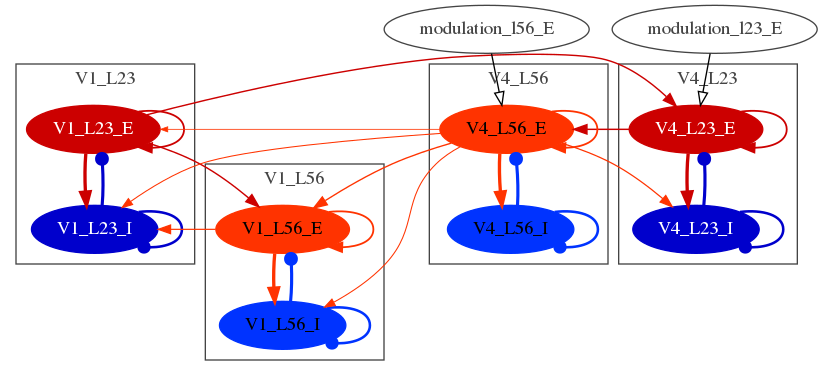
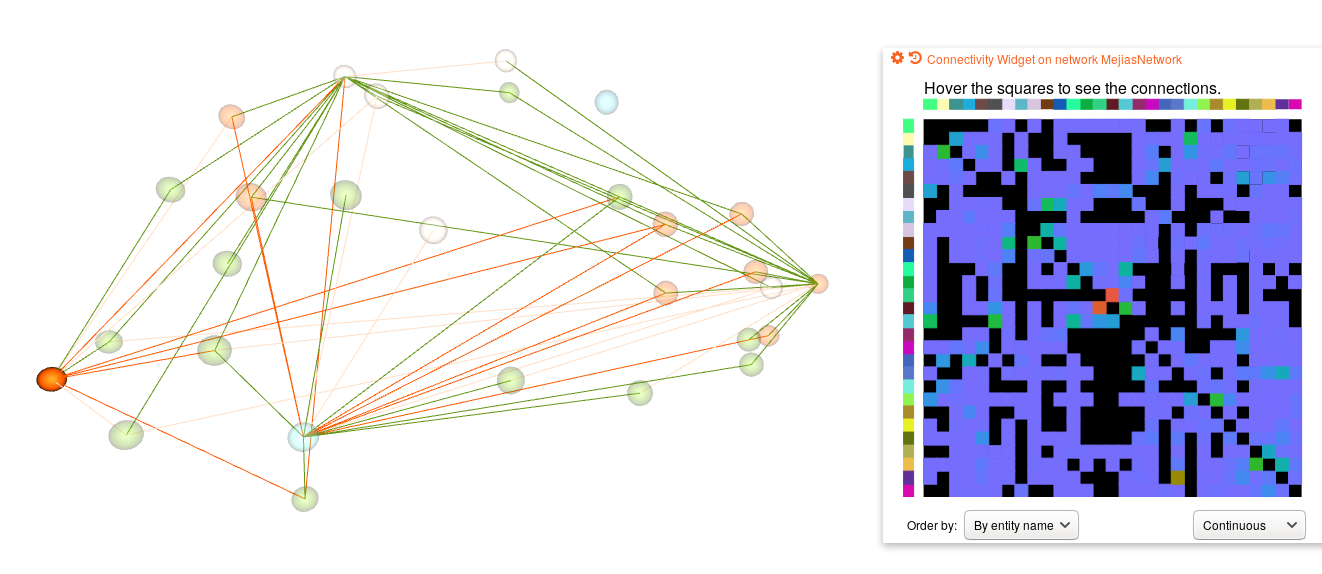
Interareal model (work in progress) View of whole cortex model on Open Source Brain (work in progress)Note: This repo is a work in progress. So far this repository contains the implementation for the model dynamics at the intralaminar and the interlaminar level. At the moment we are working on the implementation of the interareal level.
This folder contains the original model developed by Jorge Mejias and available on ModelDB.
So far, we have reproduced the main findings described by Mejias et al., 2016 at the intralaminar and interlaminar level. The main results are described here.
A basic implementation and simulation of the intralaminar model have also been implemented in NeuroML2/LEMS. GenerateNeuroMLlite.py generates the LEMS file with the description of the network.
The simulation can be run by calling inside the NeuroML2 folder:
python GenerateNeuroMLlite.py -jnml
The necessary Python packages are listed on the requirements.txt file.
As outlined above, the original Matlab scripts related to Mejias et al. 2016 were taken from ModelDB.
The data on 3D positions of areas (MERetal14_on_F99.tsv) was taken from https://scalablebrainatlas.incf.org/macaque/MERetal14#downloads; https://scalablebrainatlas.incf.org/services/regioncenters.php
-
Markov NT, Ercsey-Ravasz MM, Ribeiro Gomes AR, Lamy C, Magrou L, Vezoli J, Misery P, Falchier A, Quilodran R, Gariel MA, Sallet J, Gamanut R, Huissoud C, Clavagnier S, Giroud P, Sappey-Marinier D, Barone P, Dehay C, Toroczkai Z, Knoblauch K, Van Essen DC, Kennedy H (2014) "A weighted and directed interareal connectivity matrix for macaque cerebral cortex." Cereb Cortex 24(1):17-36.
-
Rembrandt Bakker, Paul Tiesinga, Rolf Kötter (2015) "The Scalable Brain Atlas: instant web-based access to public brain atlases and related content." Neuroinformatics. http://link.springer.com/content/pdf/10.1007/s12021-014-9258-x (arXiv)